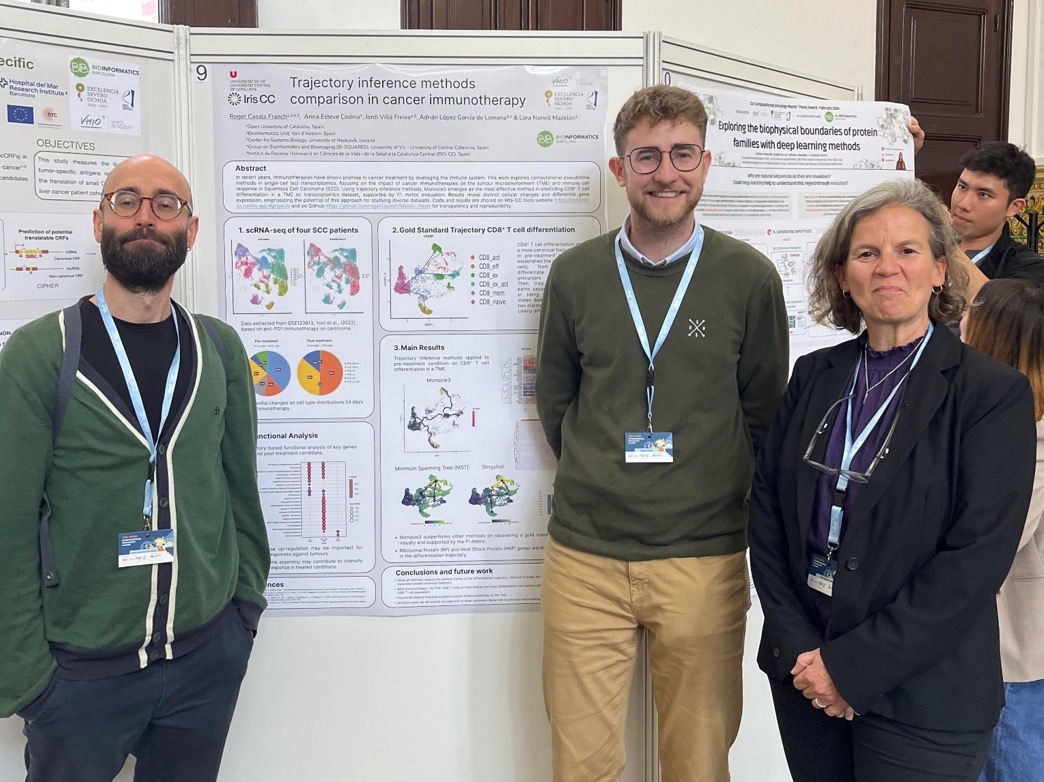
Roger Casals has participated in the first congress of the Spanish Society of Bioinformatics and Computational Biology (SEBiBC), presenting the poster entitled “Trajectory inference and transcriptional regulators of T cell response upon immunotherapy in non-melanoma skin cancer”. The congress, held at the Fira de Valencia from October 16 to 18, brought together prominent researchers in the field of bioinformatics and computational biology, consolidating itself as a success in its first edition.
In his presentation, Casals presented the results of a study that allows mapping the differentiation trajectories of T cells in the tumor microenvironment. Through a comprehensive functional analysis, the work identifies key transcription factors in the modulation of the immune response after immunotherapy in cases of non-melanoma skin cancer. These findings may be a basis for better understanding the mechanisms of response to immunotherapy, opening doors to future personalized therapies.
The work of Roger Casals has been possible thanks to the collaboration with Lara Nonell, expert in bioinformatics from the Vall d’Hebron Institute of Oncology (VHIO), as well as with the support of her doctoral co-tutors, Adrián López García de Lomana from the University of Iceland and Jordi Villà-Freixa.