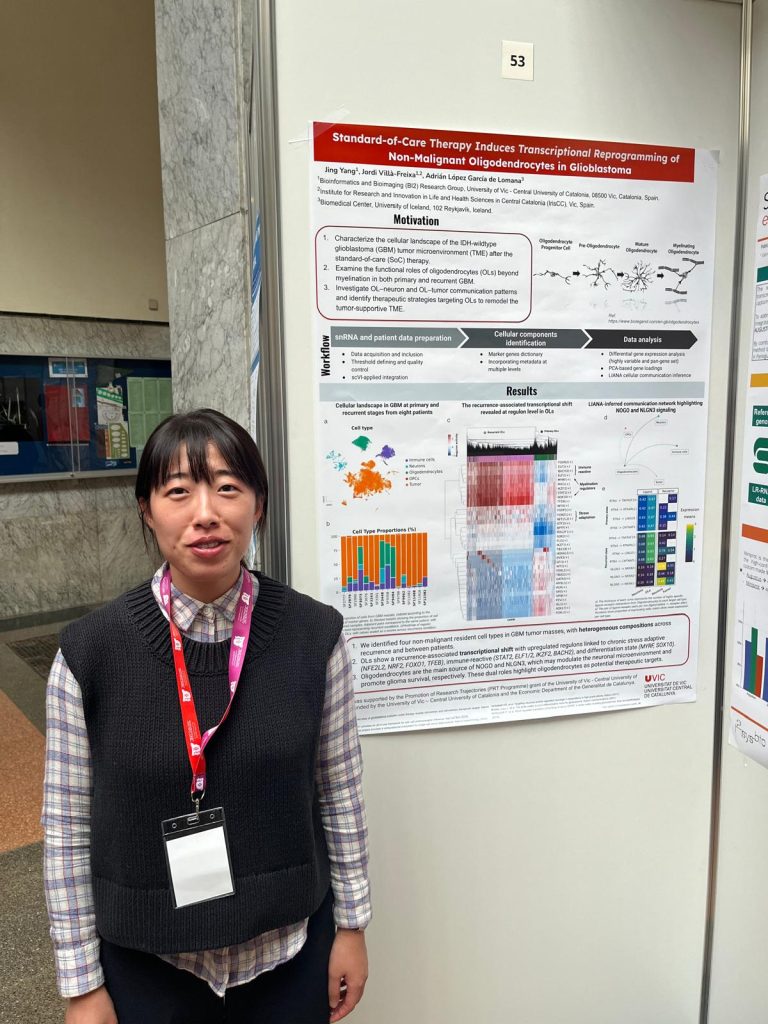
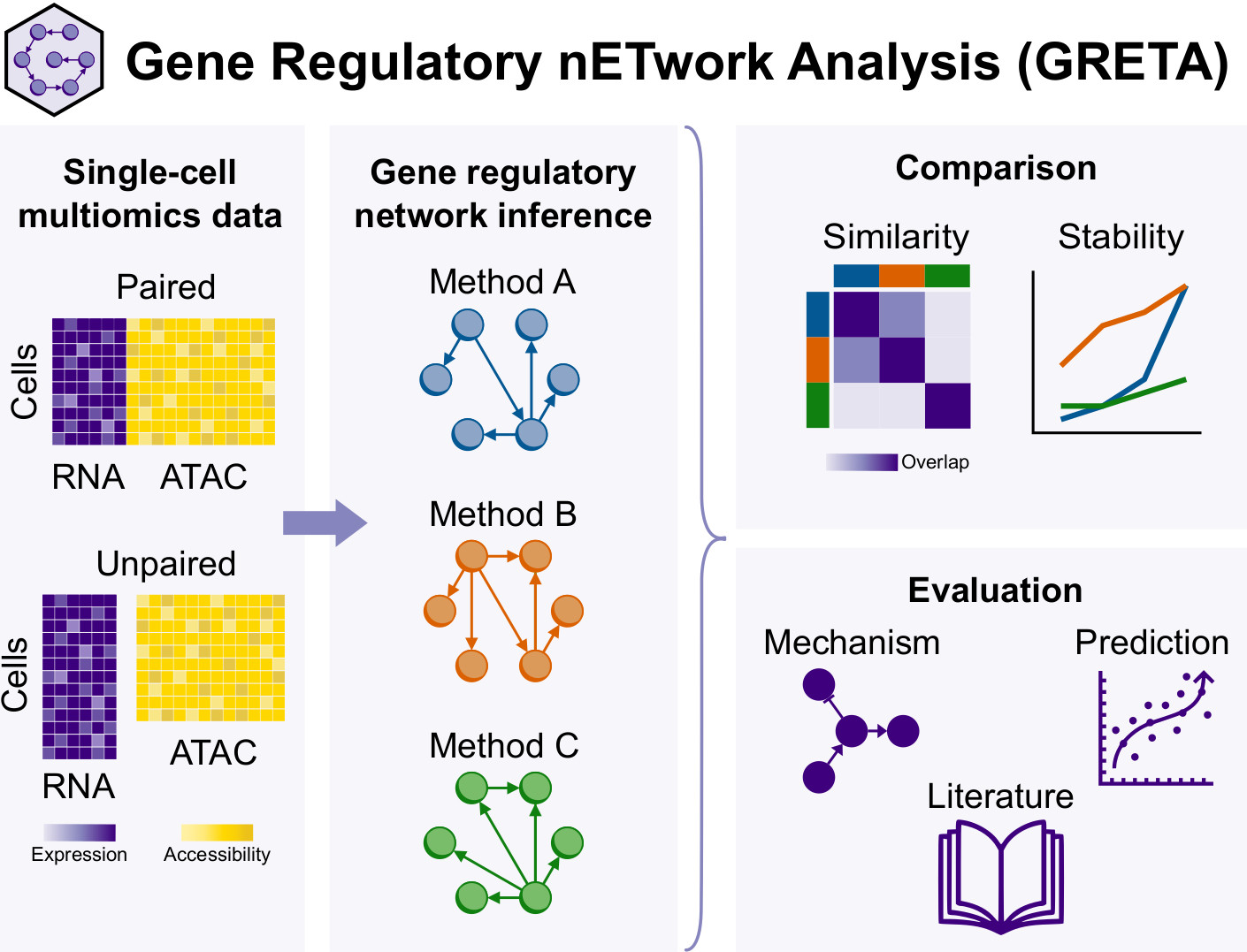
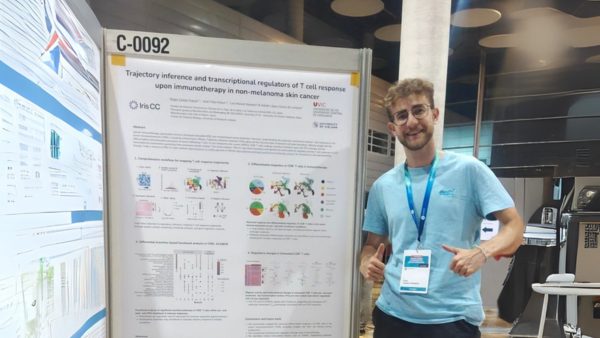
The Computational Biochemistry and Biophysics Laboratory (CBBL), part of the Bioinformatics and Bioimaging Research Group (Bi-Squared) at the University of Vic – Central University of Catalonia (UVic-UCC), in collaboration with the Central Catalonia Institute for Health Research (IRIS-CC), organized the workshop “Update in Omics Data Analysis: genome structure, metagenomics and gene regulation”. The session provided a forum to discuss and review recent developments in omics data analysis, aimed at researchers, students, and professionals in the field.
Held shortly before Christmas at the Torre dels Frares Campus in Vic, the event brought together postdoctoral researchers who presented and discussed recent advances in key areas of bioinformatics, including genome structure, metagenomics, and gene regulation using large-scale omics datasets. The workshop concluded with a round-table discussion that encouraged dialogue on emerging challenges and future directions in biological data analysis.
The program featured presentations by:
- Juan Antonio Rodríguez Pérez (University of Copenhagen) —
institutional profile,
Google Scholar - Meritxell Pujolassos (UVic-UCC) —
institutional profile,
Google Scholar - Pau Badia i Mompel (Stanford Medicine) —
institutional profile,
Google Scholar
This workshop aligns with IRIS-CC’s outreach and training initiatives and supports scientific exchange and skills development within the bioinformatics and bioimaging research community.
For more information, please visit the event webpage: Update in Omics Data Analysis: genome structure, metagenomics and gene regulation.


 and
and