In a recent article (1), the lab, including our colleague at the UVic-UCC Joan Bertran Comulada, has contributed to characterize how a well-known immune regulator, IκBα, has a surprising second role in the cell — one that could open new doors for understanding and treating inflammation, cancer, and tissue regeneration.
For decades, IκBα has been recognized as the main “off switch” of the NF-κB pathway, a critical system that controls inflammation and immune responses. Some years ago, the lab contributed to an article (2) by the team of Lluís Espinosa and coworkers at the IMIM/UPF revealing that IκBα also works inside the cell nucleus, where it interacts with DNA and chromatin to regulate genes that control stem cell identity and differentiation.
Using an innovative computational tool developed by Martin Floor while a PhD student in the CBBL, called the Fold-Excluded Evolutionary Conservation (FEEC) metric, we identified the exact regions of the protein responsible for each activity. In the study, the researchers looked not only at which parts of the protein are conserved across species, but also at how each residue fits within the protein’s 3D structure. By comparing evolutionary conservation with structural packing, they identified positions that are more conserved than expected based on their structural role. These positions may have additional regulatory or interaction functions, beyond simply maintaining the protein’s stability.
Having identified specific residues responsible for each activity prompted the creation, in this new research, of separation-of-function (SOF) mutants — engineered versions of IκBα that can perform either its traditional NF-κB–related function or its newly discovered chromatin-related one, but not both.
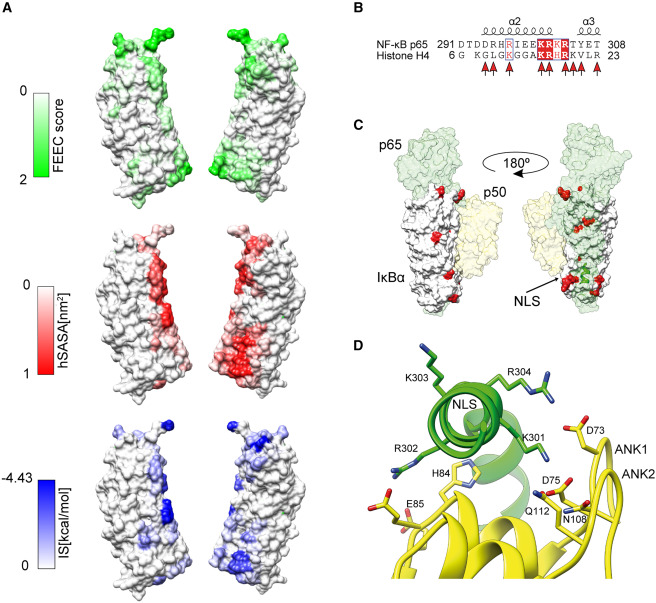
Figure 1 A common domain of IκBα is required for p65-NF-κB and histone H4 binding (1)
The subsequent experimental validation by Espinosa’s lab showed that the chromatin-associated form of IκBα is essential in intestinal stem cells, for their ability to mature into specialized cell types. This effect is independent of IκBα’s inflammatory role, highlighting its distinct influence on gene regulation and tissue health.
These findings redefine IκBα as more than just an inflammation inhibitor — it is also a key epigenetic regulator linking environmental signals to gene expression. The newly developed SOF mutants offer powerful tools to explore IκBα’s dual roles in health and disease and may guide the development of targeted therapies that fine-tune inflammation and regeneration without unwanted side effects.
1) Separation-of-function mutants reveal the NF-κB-independent involvement of IκBα in the regulation of intestinal stemness
Álvarez-Villanueva, Daniel et al.
Cell Reports, Volume 44, Issue 7, 115949, 2025
2) Chromatin-Bound IκBα Regulates a Subset of Polycomb Target Genes in Differentiation and Cancer
Mulero, María Carmen et al.
Cancer Cell, Volume 24, Issue 2, 151 – 166, 2013

 and
and