Publications
12023

Geometric deep learning reveals the spatiotemporal features of microscopic motion
Jesús Pineda, Benjamin Midtvedt, Harshith Bachimanchi, Sergio Noé, Daniel Midtvedt, Giovanni Volpe, & Carlo Manzo
Nature Machine Intelligence 5, 71-82 (2023)
[link][arXiv:2202.06355]

Preface: characterisation of physical processes from anomalous diffusion data
Carlo Manzo1, Gorka Muñoz-Gil, Giovanni Volpe, Miguel Angel Garcia-March, Maciej Lewenstein, & Ralf Metzler
Journal of Physics A: Mathematical and Theoretical 56, 010401 (2023)
[link][arXiv:2301.00800]

Broadband Plasmonic Nanoantennas for Multi-Color Nanoscale Dynamics in Living Cells
Maria Sanz-Paz, Thomas S. van Zanten, Carlo Manzo, Mathieu Mivelle, & Maria F. Garcia-Parajo
Small in press (2023)
doi: 10.1002/smll.202207977
[link]
22021
Objective comparison of methods to decode anomalous diffusion
Gorka Muñoz-Gil, Giovanni Volpe, …, Maciej Lewenstein, Ralf Metzler, and Carlo Manzo
Nature Communications 12, 6253 (2021)
[link][arXiv:2105.06766]
Extreme Learning Machine for the Characterization of Anomalous Diffusion from Single Trajectories
Carlo Manzo
Journal of Physics A: Mathematical and Theoretical 54, 334002 (2021)
[link][arXiv:2105.02597]
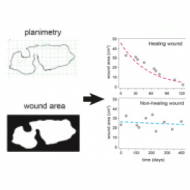
A toolkit for the quantitative evaluation of chronic wounds evolution for early detection of non-healing wounds
Marta Cullell-Dalmau, Marta Otero-Viñas, Marta Ferrer-Solà, Helena Sureda-Vidal, and Carlo Manzo
Journal of Tissue Viability 30 (2), 161-167 (2021)
doi:10.1016/j.jtv.2021.02.009
[link]
Convolutional Neural Network for Skin Lesion Classification: Understanding the Fundamentals Through Hands-On Learning
Marta Cullell-Dalmau, Sergio Noé, Marta Otero-Viñas, Ivan Meić, and Carlo Manzo
Frontiers in Medicines 8, 13 (2021)
doi:10.3389/fmed.2021.644327
[link]
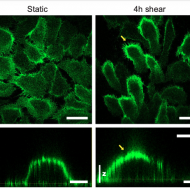
Shear forces induce ICAM-1 nanoclustering on endothelial cells that impact on T cell migration
Izabela K Piechocka, Sarah Keary, Alberto Sosa-Costa, Lukas Lau, Nitin Mohan, Jelena Stanisavljevic, Kyra JE Borgman, Melike Lakadamyali, Carlo Manzo, Maria F Garcia-Parajo
Biophysical Journal 120, 2644-2656 (2021)
doi:10.1016/j.bpj.2021.05.016
[link][biorXiv]
32020

The anomalous diffusion challenge: single trajectory characterisation as a competition
Gorka Muñoz-Gil, Giovanni Volpe, Miguel Angel García-March, Ralf Metzler, Maciej Lewenstein, and Carlo Manzo
In: Emerging Topics in Artificial Intelligence 2020 11469, 114691C (2020) – International Society for Optics and Photonics
doi:10.1117/12.2567914
[link][arXiv:2003.12036]
A machine learning method for single trajectory characterization
G. Muñoz-Gil, M. A. Garcia-March, C. Manzo, J. D. Martín-Guerrero, and M. Lewenstein
New Journal of Physics 22 (1), 013010 (2020)
doi:10.1088/1367-2630/ab6065
[link][arXiv:1903.02850]
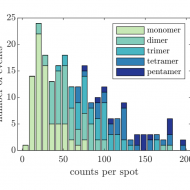
Bayesian analysis of data from segmented super-resolution images for quantifying protein clustering
T. Kosǔta, M. Cullell-Dalmau, F. Cella Zanacchi, and C. Manzo
Physical Chemistry Chemical Physics 22, 1107-1114 (2020)
doi:10.1039/C9CP05616E
[link][arXiv:1909.13133]

Research Techniques Made Simple: Deep Learning for the Classification of Dermatological Images
Marta Cullell-Dalmau, Marta Otero-Viñas, and Carlo Manzo
Journal of Investigative Dermatology 140 (3), 507-514.e1 (2020)
doi: 10.1016/j.jid.2019.12.029
[link]

Dynamic actin-mediated nano-scale clustering of CD44 regulates its meso-scale organization at the plasma membrane
Parijat Sil, Nicolas Mateos, Sangeeta Nath, Sonja Buschow, Carlo Manzo, Kenichi G. N. Suzuki, Takahiro Fujiwara, Akihiro Kusumi, Maria F. Garcia-Parajo, and Satyajit Mayor
Molecular Biology of the Cell 31 (7), 561-579 (2020)
doi: 10.1091/mbc.E18-11-0715
[link]
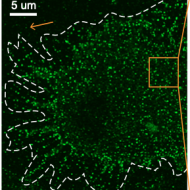
Membrane receptor MerTK is a newly identified transcriptional regulator that associates to chromatin as nanoclusters during human DC differentiation
Kyra JE Borgman, Georgina Flórez-Grau, Maria A Ricci, Carlo Manzo, Melike Lakadamyali, Alessandra Cambi, Daniel Benítez-Ribas, Felix Campelo, Maria F Garcia-Parajo
biorXiv (2020)
doi:10.1101/2020.04.16.044974
[biorXiv]
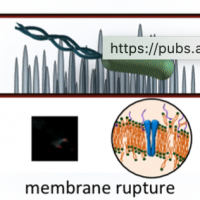
Direct and fast assessment of antimicrobial surface activity using molecular dynamics simulation and time-lapse imaging
Rafaël Sibilo, Ilaria Mannelli, Ramon Reigada, Carlo Manzo, Mehmet A Noyan, Prantik Mazumder, and Valerio Pruneri
Analytical Chemistry 92 (10), 6795-6800 (2020)
doi: 10.1021/acs.analchem.0c00367
[link]
42019
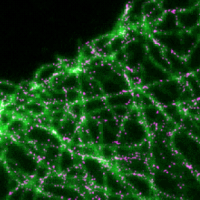
Quantifying motor protein copy number in super-resolution using an imaging invariant calibration
F. Cella Zanacchi, C. Manzo, R. Magrassi, N.D. Derr, M. Lakadamyali
Biophysical Journal 116, 2195-2203 (2019) [link]
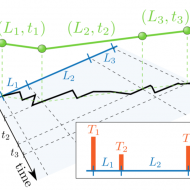
Diffusion through a network of compartments separated by partially-transmitting boundaries
G. Muñoz-Gil, M.A. García-March, C. Manzo, A. Celi, M. Lewenstein
Frontiers in Physics 7, 31 (2019) [link]
doi: 10.3389/fphy.2019.00031
Inhomogeneous membrane receptor diffusion explained by a fractional heteroscedastic time series model
M. Balcerek, H. Loch-Olszewska, J. A. Torreño-Piña, M. F. Garcia-Parajo, A. Weron, C. Manzo, K. Burnecki
Physical Chemistry Chemical Physics 21, 3114-3121 (2019) [link]
52018
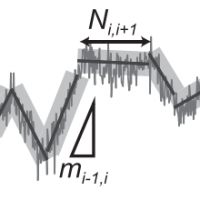
PLANT: A method for detecting changes of slope in noisy trajectories
A. Sosa-Costa, I. K. Piechocka, L. Gardini, F. S. Pavone, M. Capitanio, M. F. Garcia-Parajo, C. Manzo
Biophysical Journal 114, 2044-2051 (2018) [link]
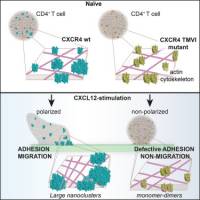
Separating actin-dependent chemokine receptor nanoclustering from dimerization indicates a role for clustering in CXCR4 signaling and function
L. Martínez-Muñoz, J. . Rodríguez-Frade, R. Barroso, C. O. S. Sorzano, J. A. Torreño-Pina, C. A. Santiago, C. Manzo, P. Lucas, E. M. García-Cuesta, E. Gutierrez, L. Barrio, J. Vargas, G. Cassio, Y. R. Carrasco, F. Sánchez-Madrid, M. F. García-Parajo, M. Mellado
Molecular Cell 70, 106-119 (2018) [link]
62017
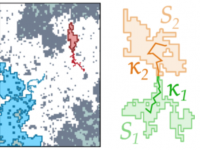
Transient subdiffusion from an Ising environment
G. Muñoz-Gil, C. Charalambous, M. A. García-March, M. F. Garcia-Parajo, C. Manzo, M. Lewenstein, and A. Celi
Physical Review E 96, 052140 (2017) [link]
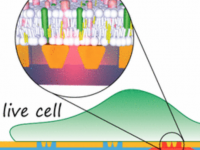
Planar optical nanoantennas resolve cholesterol-dependent nanoscale heterogeneities in the plasma membrane of living cells
R. Regmi , P. M. Winkler , V. Flauraud , K. J. E. Borgman, C. Manzo, J. Brugger , H. Rigneault, J. Wenger, M. F. García-Parajo
Nano Letters 17, 6295-6302 (2017) [link]
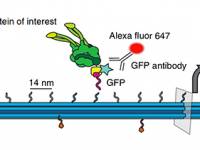
A DNA origami platform for quantifying protein copy number in super-resolution
F. Cella Zanacchi, C. Manzo, A. S. Alvarez, N. D. Derr, M. F. Garcia-Parajo, M. Lakadamyali
Nature Methods 14, 789-792 (2017) [link]

Nonergodic subdiffusion from transient interactions with heterogeneous partners
C. Charalambous, G. Muñoz-Gil, A. Celi, M. F. Garcia-Parajo, M. Lewenstein, C. Manzo, M. A. García-March
Physical Review E 95, 032403 (2017) [link]
72016
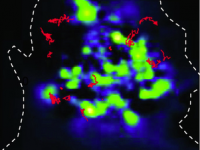
Changes in membrane sphingolipid composition modulate dynamics and adhesion of integrin nanoclusters
C. Eich, C. Manzo, S. de Keijzer, G.-J. Bakker, I. Reinieren-Beeren, M. F. Garcia-Parajo, A. Cambi
Scientific Reports 6, 20693 (2016) [link]

Uncovering homo-and hetero-interactions on the cell membrane using single particle tracking approaches
J. A. Torreno-Pina, C. Manzo, M. F. Garcia-Parajo
Journal of Physics D: Applied Physics 49 (10), 104002 (2016) [link]